Months after the first SARS-CoV-2 variant has been sequenced, many scientific works have largely hypothesized the clinical impact of the mutations, based on in silico experiments.
Starting point
The role of SARS-CoV-2 mutations has been supposed to impact the immune response strength, letting them able to escape from neutralizing – vaccines and natural infection-induced – antibodies (Fratev 2020; Teng et al. 2021), and boosting viral transmissibility.
Concerns
Major concerns linked to variants are associated with the recently identified SARS-CoV-2 strains that have acquired mutations in the ACE2 interaction region of Spike/Receptor Binding Domain, such as N501Y in B.1.1.7 (aka UK variant) and K417N, E484K, and N501Y in B.1.351 (aka South Africa variant). These SARS-CoV-2 variants, thanks to their mutation, have seen the fastest spread until becoming the dominant strains first in the regions where they were first identified, and then globally. B.1.1.7 and B.1.351 are striking examples of what it means, as largely demonstrated by data collected about B.1.1.7 spread in the UK..
The first experimental confirmation of the major concerns about the south africa variant.
A very recent study published on Cell (Zhou et al. 2021) and named: “Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera”, demonstrated as mutations responsible for B.1.351 V.O.C. (aka south African variant), confer virus resistance when exposed to immune serum large cohort of convalescent or vaccinee serum samples originated from non-mutated strains.
During the experimentation, researchers analysed:
- Neutralization of B.1.351 by convalescent plasma
- Neutralization of B.1.351 by vaccinee serum
- Neutralization of B.1.351 by a large panel of mAbs
- Neutralization of B.1.351 by monoclonal antibodies (mAbs) in late-stage clinical trials
It has been demonstrated that B.1.351 SARS-CoV-2 strain is much more difficult to neutralize than parental strains; 14 of 20 of a panel of mAbs appeared knocked out.
The cause of the widespread escape from monoclonal antibody neutralization is largely driven by E484K mutation, although K417N and N501Y act together against some important antibody classes.
On convalescent serum, the neutralization titers are reduced more than 13-fold for B.1.351 compared with the standard SARS-CoV-2 strain.
Neutralization titers for the Oxford-Astrazeneca and Pfizer vaccines
Neutralization titers for the Oxford-AstraZeneca and Pfizer vaccines (serum) were similarly reduced with B.1.351 by 9-fold and 7.6-fold, respectively.
In a number of cases, it would appear that convalescent and some vaccine serum offers limited protection against this variant (Zhou et al. 2021).
The good news
The good news is that neutralizing antibodies generated in patients with South Africa variant infections showed high levels of cross-reactivity for the original variant (Moyo-Gwete et al. 2021).
What worth a deepest look into
The authors finish their work with an important piece of advice: “Intensive surveillance systems need to be implemented to monitor for the emergence of new variants and, in particular, to be targeted at searching for breakthrough infections in vaccinees. Work on second and even third-generation vaccines to target variant viruses and more broadly to develop immunogens to targets less reliant on the ACE2-RBD interaction surface are deserving of further study”.
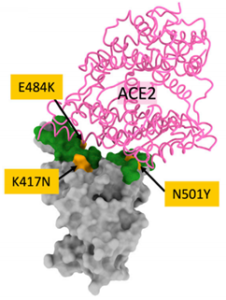
Positions of major changes in South Africa variant’s spike protein are highlighted and tagged within the Receptor Binding Domain (RBD- green). ACE2 receptor in pink. (Zhou et al. 2021).
Written by Simone Di Giacomo, PhD, OaCP R&D Manager

CoViD-19 Nucleic Acid Detection Kit (VARIANTS)
Bibliography:
Fratev, Filip. 2020. “The N501Y and K417N Mutations in the Spike Protein of SARS-CoV-2 Alter the Interactions with Both HACE2 and Human Derived Antibody: A Free Energy of Perturbation Study.” bioRxiv: 2020.12.23.424283. http://biorxiv.org/content/early/2020/12/31/2020.12.23.424283.abstract .
Moyo-Gwete, Thandeka et al. 2021. “SARS-CoV-2 501Y.V2 (B.1.351) Elicits Cross-Reactive Neutralizing Antibodies.” bioRxiv : the preprint server for biology.
Teng, Shaolei et al. 2021. “Systemic Effects of Missense Mutations on SARS-CoV-2 Spike Glycoprotein Stability and Receptor-Binding Affinity.” Briefings in bioinformatics.
Zhou, Daming et al. 2021. “Evidence of Escape of SARS-CoV-2 Variant B.1.351 from Natural and Vaccine-Induced Sera.” Cell.




